https://www.genzentrum.uni-muenchen.de/research-groups/beckmann/index.html
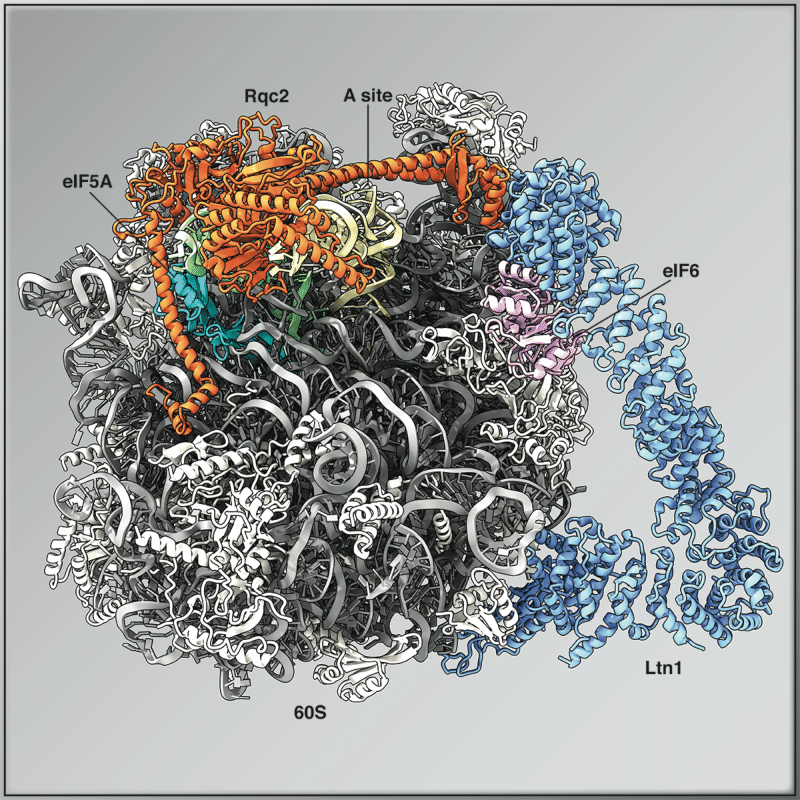
Roland Beckmann’s lab is interested in the decoding of the molecular mechanisms underlying protein biosynthesis, its regulation and ribosome-associated activities (Fig. 1), and of eukaryotic ribosome biogenesis (Fig. 2). The central technology used is cryo-electron microscopy (cryo-EM) to determine the structure of functionally defined complexes involved in these processes. In addition, complementary molecular biological and biochemical methods are used, in particular so-called cell-free systems, to further characterize the functioning of these complexes. A long-term goal is to use these findings as a basis to intervene in or repurpose both, protein production and ribosome biogenesis, in order to develop novel macromolecules and nanomachines. A current focus is the reconstitution, analysis and manipulation of the so-called CAT tailing system (Fig. 3) which allows for 40S-, mRNA- and GTPase-independent peptide elongation during ribosomal quality control (RQC).
References
- P. Tesina*, S. Ebine*, R. Buschauer*, M. Thoms, Y. Matsuo, T. Inada, R. Beckmann, Molecular Cell 2023, accepted
- M. Ameismeier, I. Zemp, J. van den Heuvel, M. Thoms, O. Berninghausen, U. Kutay, R. Beckmann, Structural basis for the final steps of human 40S ribosome maturation. Nature 2020 , doi: 10.1038/s41586-020-2929-x
- J. Cheng, B. Lau, G. La Venuta, M. Ameismeier, O. Berninghausen, E. Hurt, R. Beckmann, 90S pre-ribosome transformation into the primordial 40S subunit. Science 2020 , 369(6510):1470-1476, doi: 10.1126/science.abb4119
- M. Thoms, R. Buschauer, M. Ameismeier, L. Koepke, T. Denk, M. Hirschenberger, H. Kratzat, M. Hayn, T. Mackens-Kiani, J. Cheng, J. H. Straub, C. M. Stürzel, T. Fröhlich, O. Berninghausen, T. Becker, F. Kirchhoff, K. Sparrer, R. Beckmann, Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, doi: 10.1126/science.abc8665
- R. Buschauer, Y. Matsuo, T. Sugiyama, Y. H. Chen, N. Alhusaini, T. Sweet, K. Ikeuchi, J. Cheng, Y. Matsuki, R. Nobuta, A. Gilmozzi, O. Berninghausen, P. Tesina, T. Becker, J. Coller, T. Inada, R. Beckmann, The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020 , 368(6488) pii: eaay6912, doi: 10.1126/science.aay6912
- T. Su, T. Izawa, M. Thoms, Y. Yamashita, J. Cheng, O. Berninghausen, F. U. Hartl, T. Inada, W. Neupert, R. Beckmann, Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis. Nature 2019 , doi: 10.1038/s41586-019-1307-z

