The group of Protein Design and Self-Assembly focuses on dissecting the physical principles of protein self-assembly. While conceptually simple, the phenomenon of self-assembly entails a fine equilibrium of a number of physical properties, which determine the dynamics and structure of the assembly architecture. We combine computational de novo protein design with protein production and in vitro biophysical methods to systematically investigate the interplay between different types of interactions in the protein assembly process. Our goal is to create dynamic and responsive protein-based materials that can interface with biological systems.
References
-
Jing Yang (John) Wang∗, Alena Khmelinskaia∗, et al. Improving the secretion of
designed protein assemblies through negative design of cryptic transmembrane domains. PNAS, 120 (11) e2214556120. https://www.pnas.org/doi/10.1073/pnas.2214556120 -
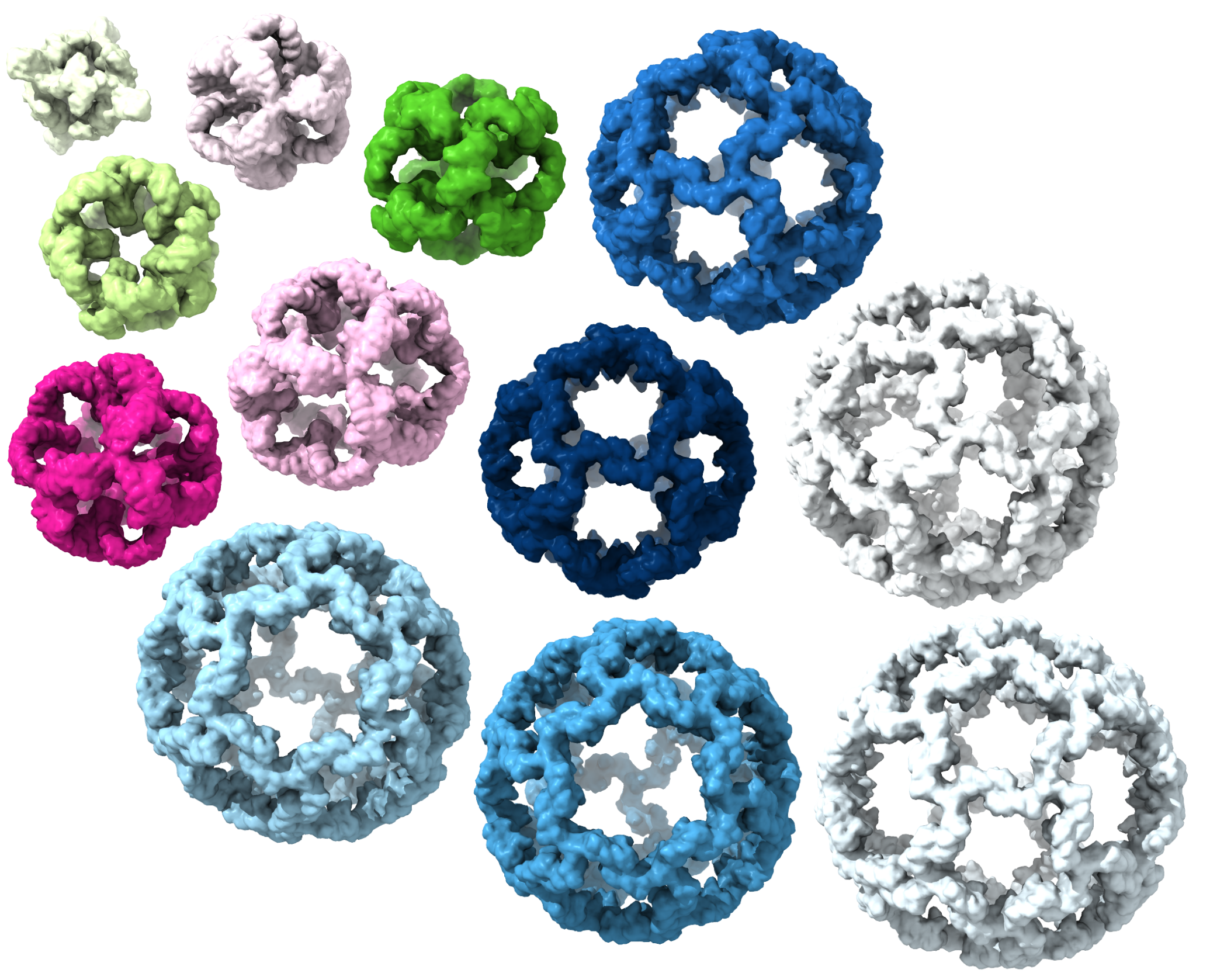
Bhoomika Basu Mallik∗, Jenna Stanislaw∗, Tharindu Madhusankha Alawathurage∗, Alena Khmelinskaia. De novo design of polyhedral protein
assemblies: before and after the AI revolution. ChemBioChem, doi: 10.1002/cbic.202300117 https://doi.org/10.1002/cbic.202300117 -
Will Sheffler∗, Erin C. Yang∗, Quinton Dowling∗, Yang Hsia∗, Chelsea N. Fries, Jenna Stanislaw, Mark D. Langowski, Marisa Brandy, Alena Khmelinskaia, Neil P. King, David A. Baker. Fast and versatile sequence-independent protein docking for nanomaterials design using RPXDock. PLoS Comput Biol 19(5): e1010680. https://doi.org/10.1371/journal.pcbi.1010680
-
A Khmelinskaia#, NP Bethel*, F Fatehi*, A Antanasijevic, AJ Borst, S-H Lai, JY(J) Wang, B Basu Mallik, MM Miranda, AM Watkins, C Ogohara, S Caldwell, M Wu, AJR Heck, D Veesler, AB Ward, D Baker, R Twarock, NP King# (2023) Local structural flexibility drives oligomorphism in computationally designed polyhedral protein assemblies. bioRxiv doi: 10.1101/2023.10.18.562842
- Erin C. Yang, Robby Divine, Marcos C. Miranda, Andrew J. Borst, Will Sheffler, Jason Z. Zhang, Justin Decarreau, Amijai Saragovi, Mohamad Abedi, Nicolas Goldbach, Maggie Ahlrichs, Craig Dobbins, Alexis Hand, Suna Cheng, Mila Lamb, Paul M. Levine, Sidney Chan, Rebecca Skotheim, Jorge Fallas, George Ueda, Joshua Lubner, Masaharu Somiya, Alena Khmelinskaia, Neil P. King, David Baker. Computational design of non-porous, pH-responsive antibody nanoparticles. Nat Struct Mol Biol

