https://www.softmatter.physik.uni-muenchen.de/raedler_group/index.html

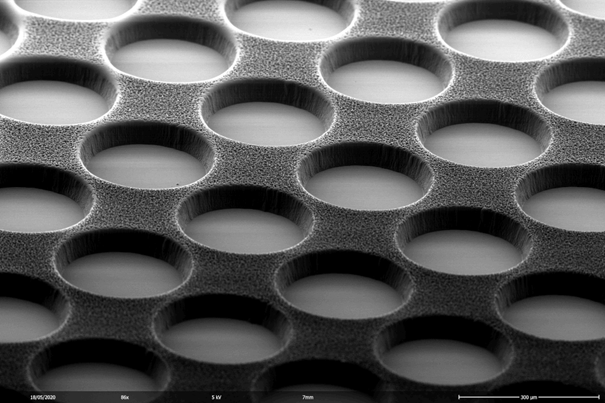

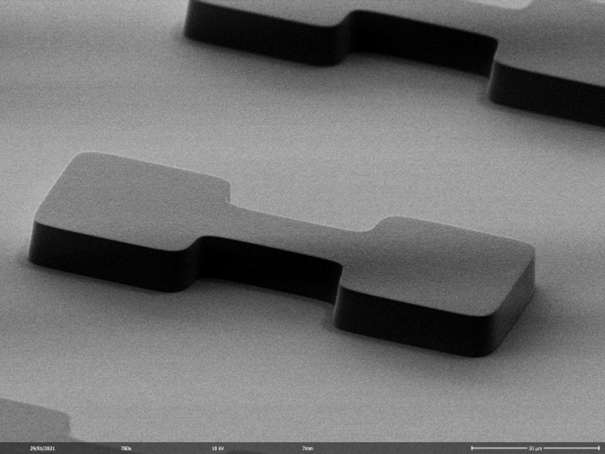
We fabricate artificially designed micro-environments to culture and probe living cells. Arrays of defined cavities provide standardized geometric boundaries for individual cells or groups of cells. Imaging such cell-arrays in scanning time lapse mode yields hundreds of single cell trajectories and hence ample statistics for computational data analysis. We study single cell gene expression, cell migration and apoptotic events with precise spatio-temporal control. To this end we develop biofabrication techniques, including photo-lithography, hot-embossing, galvanic, soft lithography, 3D printing and microcontact printing, to create two-dimensional (2D) and three-dimensional (3D) pattern, cavities and channels made of biocompatible materials like PDMS, COC, hydrogels and proteins.The assays allow for long-term observation of growth and function of individual living cells, interacting groups of cells and organoids. We aim to build 3D microenvironments that control cell behavior, such as guidance of cell migration and guided interaction with nanoscale objects including cell-lipidnanoparticle interactions.
References
- A. Reiser, D. Woschée, S. M. Kempe, J. O. Rädler, Live-cell Imaging of Single-Cell Arrays (LISCA) - a Versatile Technique to Quantify Cellular Kinetics. J. Vis. Exp. 2021, (169), e62025
- C. Schreiber, B. Amiri, J. C. J. Heyn, J. O. Rädler, M. Falcke, On the adhesion–velocity relation and length adaptation of motile cells on stepped fibronectin lanes. PNAS 2021, 118 (4)
- B. Brückner, A. Fink, C. Schreiber, P. J. F. Röttgermann, J. O. Rädler, and C. P. Broedersz, Stochastic Nonlinear Dynamics of Confined Cell Migration in Two-State Systems. Nature Physics 2019, 19, 1592
- M. Dietrich, H. Le Roy, D. B. Brückner, H. Engelke, R. Zantl, J. O. Rädler, and C. P. Broedersz, Guiding 3D Cell Migration in Deformed Synthetic Hydrogel Microstructures. Soft Matter 2018, 15
- C. Schreiber, F. J. Segerer, E. Wagner, A. Roidl & J. O. Rädler, Ring-Shaped Microlanes and Chemical Barriers as a Platform for Probing Single-Cell Migration. Scientific Reports 2016
- I. Chatzopoulou, C. C. Roskopf, F. Sekhavati, T. A. Braciak, N. C. Fenn, K.-P. Hopfner, F. S. Oduncu, G. H. Fey, J. O. Rädler, Chip-based platform for dynamic analysis of NK cell cytolysis mediated by a triplebody. Analysis 2016 , Advance Article, DOI:10.1039/C5AN02585K
- J. Segerer, P. J. F. Röttgermann, S. Schuster, A. P. Alberola, S. Zahler, J. O. Rädler, Versatile method to generate multiple types of micropatterns. Biointerphases 2016 , 11, 011005
- M. Ferizi, C. Leonhardt, C. Meggle, M. K. Aneja, C. Rudolph, C. Plank, J. O. Rädler, Stability analysis of chemically modified mRNA using micropattern-based single-cell arrays. Lab on a Chip 2015 , DOI: 10.1039/c5lc00749f

